Research overview
Our main research line focuses on the interaction of ionic liquids (ILs) - a vast and relatively new class of organic electrolytes - with the basic building blocks of living matter, including lipid bilayers and biomembranes, proteins and amyloids, and cells, with the aim to understand the chemical-physical properties relevant to the development of applications in pharmacology, nano-bio medicine and nano-bio technology. Neutron scattering, atomic force microscopy and computer simulations are the three major techniques used for these investigations, which are combined with a number of complementary approaches, including cell-biology assays.
Our second line of research is aimed at the development of a new neutron scattering method/spectroscopy for dynamics. This novel approach is based on the detection of the solely neutrons that are scattered into the elastic line and offers several advantages in comparison to standards methods for dynamics, which make it particularly suitable for the study of biomolecules. Theoretical approaches and Monte Carlo computations are the two methods employed here, which are completed by test experiments at large-scale neutron scattering facilities.
Our third research line focuses on water structure and dynamics at (bio)-interfaces and in confinement, with a special focus on water molecules confined at biological surfaces. Other interests include the investigation of the dynamics-function relationship in proteins. Neutron scattering and classical molecular dynamics simulations are the two major approaches adopted in these studies.
Methods:
Atomic force microscopy, neutron scattering, and computer simulations are the three major techniques in use in our Lab with the aim to link molecular, if not, atomistic-scale mechanisms to behaviours and processes taking place at the meso-scale (Fig. 1). A series of complementary approaches are also used, which include static and dynamic light scattering, calorimetry, optical and electron microscopies, Raman and infrared spectroscopies and a palette of different biochemical and cell biology methods as, for example, cell survival assays, western blotting of key proteins, flow cytometry, cell migration and scattering assays.
Atomic force microscopy, neutron scattering, and computer simulations are the three major techniques in use in our Lab with the aim to link molecular, if not, atomistic-scale mechanisms to behaviours and processes taking place at the meso-scale (Fig. 1). A series of complementary approaches are also used, which include static and dynamic light scattering, calorimetry, optical and electron microscopies, Raman and infrared spectroscopies and a palette of different biochemical and cell biology methods as, for example, cell survival assays, western blotting of key proteins, flow cytometry, cell migration and scattering assays.
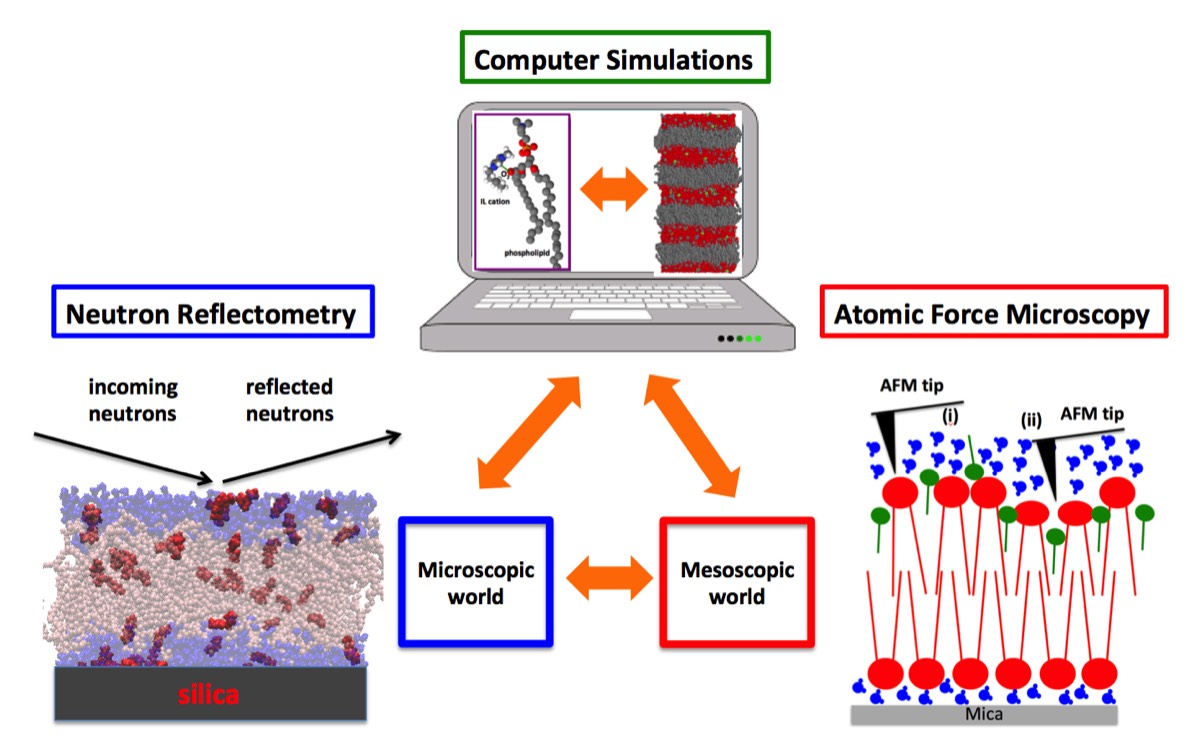
Fig. 1 - A representative example. Neutron reflectometry allowed to determine the partitioning of ionic liquid (IL) cations between lipid and aqueous phases of a model biomembrane, which quantitatively agreed with the classical molecular dynamics (MD) simulations carried out on the same system. The analysis of the MD trajectories suggested that IL-cations alter the viscoelasticity of the membrane, which has been investigated then experimentally by atomic force microscopy.